Detailed view of viral replication machinery lends new insights into infection
The coronavirus that causes COVID-19, SARS-CoV-2, is known as a positive-strand RNA virus because of the way it stores and makes copies of its genetic material. Many other important pathogens such as the Zika, dengue and chikungunya viruses are also part of this same group — the largest of six genetic classes of viruses.
Scientists at the Morgridge Institute for Research and the University of Wisconsin–Madison have captured strikingly improved images of the machinery these viruses use to make copies of, or replicate, their RNA genomes. The findings should aid researchers developing antiviral drugs and treatments, and provide new insights into how this large class of viruses work.
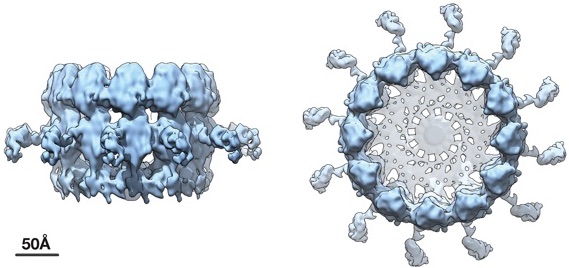
Cryoelectron microscope tomography imaging reveals high-resolution side and top views of the viral RNA replication “crown” complex structure. Courtesy of Paul Ahlquist
In 2017, a team led by Paul Ahlquist, director of the John W. and Jeanne M. Rowe Center for Research in Virology at the Morgridge Institute and professor of oncology and molecular virology at UW–Madison, first revealed the existence of this crown-like viral RNA replication complex. The team showed that the so-called crowns contained proteins the viruses use to read and copy RNA.
The new research led by Ahlquist and published July 20, 2020, in the Proceedings of the National Academy of Sciences builds upon that work by using an advanced technique called cryoelectron microscope (cryo-EM) tomography to dramatically improve the resolution of images of the replication crown complex.
“Cryo-EM has recently gone through a quantum leap in its capabilities,” Ahlquist says. “In this study our research group combined multiple advances to greatly improve sample preparation, image acquisition and image processing, and to map the position of specific protein domains in the complex.”
Within an infected cell, viral RNA replication happens at cellular membranes, often upon the formation of virus-induced compartments known as spherules. Ahlquist and his team previously showed that the spherules contain and protect a copy of the viral RNA genome, which functions as a template from which additional copies are made.
The replication complex repeatedly copies this archival viral RNA to produce new genomes that are released from the spherule through a membranous neck and then incorporated as the payload of new infectious virions.
In each positive-strand RNA virus, most of the viral genes are devoted to this single process of RNA reproduction.
“Given this massive investment of resources, viral RNA genome replication is arguably one of the most important processes in infection, and it is already a major target for virus control,” Ahlquist says.
The team’s prior work also showed that the crown structure, which sits atop the spherule neck, contains a key viral protein that creates the replication complex and copies the viral RNA.
The new higher-resolution cryo-EM images and complementary results show that the crown is composed of 12 copies of the key viral RNA replication protein, arranged like staves in a barrel.
It also provides the team with clues as to how it works. The replication protein is large and contains segments, or domains, that perform different functions. Two of these domains — RNA polymerase and RNA capping domains— are common across numerous positive-strand RNA viruses and contribute to synthesizing new viral genome copies.
The findings should aid researchers developing antiviral drugs and treatments, and provide new insights into how this large class of viruses work.
Using an approach that involved labeling part of the replication complex with gold nanoparticles, Ahlquist’s team discerned the orientation of these domains within the crown structure. How these domains are physically organized is one of the most important issues for understanding how the replication complex functions.
For instance, they found that the polymerase domain that actually synthesizes new RNA is positioned at the apex of the crown, providing clues about the early steps in the replication process in which the starting viral RNA template is recruited into the complex and the spherule is formed. It also provides context for later steps, during which the template is copied to make progeny genomes for packaging into new infectious virus particles.
“Such advances will reveal in increasing detail how these complexes assemble and operate, and thus how they might be best attacked,” Ahlquist explains. “These insights should provide the basis for novel, stronger antiviral mechanisms.”
The results also provide a strong foundation for further experiments to define the replication complex structure and function at even higher levels, the researchers say.
“We hope to continue to improve the RNA replication complex crown structure to provide additional important refinements in the future,” Ahlquist says. “We also hope to address growing indications from our work that conformational changes in these proteins are critical to their multiple functions.”
A portion of the study was supported by NIH Grant U24GM129547 and performed at the Pacific Northwest Center for Cryo-EM at Oregon Health & Science University and accessed through EMSL (grid.436923.9), a Department of Energy Office of Science User Facility sponsored by the Office of Biological and Environmental Research.
This research was performed using computing resources and assistance from the UW–Madison Center for High Throughput Computing in the Department of Computer Sciences, which is supported by UW–Madison, the Advanced Computing Initiative, the Wisconsin Alumni Research Foundation, the Wisconsin Institute for Discovery, and the National Science Foundation, and is an active member of the Open Science Grid, which is supported by the NSF and the U.S. Department of Energy’s Office of Science.
Ahlquist is an investigator of the Howard Hughes Medical Institute and the Morgridge Institute for Research, from which he received support. He also received support from the NIH and the John W. and Jeanne M. Rowe Center for Research in Virology.