Breaking up (protein complexes) is hard to do, but new UW study shows how
A new study by University of Wisconsin–Madison researchers identified the structural basis for how tightly bound protein complexes are broken apart to become inactivated. The structure explains why the complexes are less active in some cancers and neurodegenerative diseases, and offers a starting point to identify drug targets to reactivate it.
As we grow, our cells respond to tightly regulated cues that tell them to grow and divide until they need to develop into specialized tissues and organs. Most adult cells are specialized, and they correctly respond to cues that tell them to stop growing. Cancers can develop when something goes awry with those cues.
One such “stop and specialize” cue is found with the protein complexes known as PP2A. There are approximately 100 known PP2A complexes, and together they are estimated to regulate nearly one-third of all cellular proteins. These complexes consist of a core that is inactive until it mixes and matches with one of several specificity proteins to form tightly bound, active PP2A complexes. Active PP2A uses those specificity partners to find its targets – typically pro-growth proteins – and inactivates them. PP2A is a critical cue, then, in keeping cell growth in check and maintaining normal neurological functions. Not surprisingly, it is mutated in many cancers and neurological disorders.
“We know a lot about how active PP2A complexes form and are identifying more and more of their targets in cells, but we know very little about how they are inactivated,” explains Yongna Xing, an associate professor of oncology with the UW Carbone Cancer Center and McArdle Laboratory for Cancer Research and the senior author of a new study published today (Dec. 22, 2017) in Nature Communications. “It’s a very tightly held complex, it’s almost like a rock, but there has to be a way to break it up.”
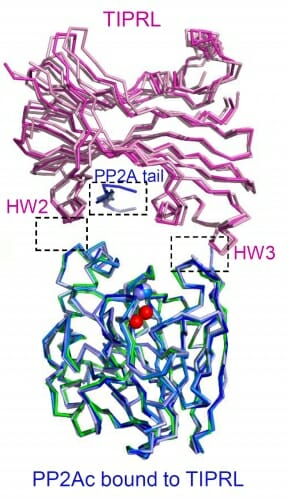 Xing’s previous work showed that PP2A is inactive when a regulatory protein, a4, is attached. However, when active PP2A complexes were challenged with a4, they remained active, meaning there had to be another trigger that broke the complex apart.
Xing’s previous work showed that PP2A is inactive when a regulatory protein, a4, is attached. However, when active PP2A complexes were challenged with a4, they remained active, meaning there had to be another trigger that broke the complex apart.
In the new study, Xing and her colleagues identify that trigger as the protein TIPRL. When they challenged active PP2A complexes with a4 and TIPRL, the complexes broke apart. Next, they determined the three-dimensional structure of TIRPL with PP2A through a technique known as X-ray crystallography.
“The structure shows how TIPRL can attack active PP2A complexes even though it has a much lower affinity than the specificity subunits do for PP2A core,” Xing says. “With the structure we were able to identify how TIRPL can attack the complex, change its conformation and, together with a4, make it fall apart robustly. It was hard to picture how this process could happen without structural insights.”
If we think of PP2A as a power screwdriver, the findings make a lot of practical sense. The core protein is the motorized base, and the specificity proteins – the ones that mix and match to help PP2A find the right target – are the screw heads. When you want to switch from a Phillips-head to a flathead screwdriver, you don’t throw away the whole power screwdriver complex and buy a new one; rather you detach one screw head and attach another. Similarly, it is energy costly for a cell to degrade the entire PP2A complex, so TIPRL’s role is to detach the specificity protein and recycle PP2A core.
One of the more interesting findings from the structure was how flexible TIRPL is compared to the specificity subunits, prompting the researchers to ask how PP2A mutations commonly seen in cancer patients affect TIPRL binding. Using either normal or PP2A core containing these mutations, they measured how well TIPRL and the specificity subunits can bind to it. They found that the core mutations have almost no effect on TIPRL binding, but they drastically weaken the binding of specificity proteins. These mutations, then, likely cause a shift from active PP2A complexes to the disassembled and inactive form.
“In many diseases, including cancers and neurodegenerative diseases, PP2A in general is less active, often due to mutations,” Xing notes. “This structure helps explain how those mutations lead to downregulation of PP2A by shifting the balance toward TIPRL-induced complex dissociation.”
With the structure in hand, Xing expects to be able to better understand the cycle of PP2A activation and inactivation, and how it regulates cell growth.
“For example, active PP2A is known to inhibit K-ras, a protein that drives growth in many tumors and currently has no good clinical inhibitors,” Xing says. “If you can find a way to re-activate PP2A, it could be very important in treating those cancers.”
The study was supported by National Institutes of Health grant R01 GM096060-01.




